Preparation of the template DNA
The design and preparation of template DNA for PUREfrex® is a very important step for successful protein synthesis. The following is a summary of the points for the preparation of template DNA.
CONTENTS
Template DNA for PUREfrex®
Both circular and linear DNA (including PCR product and circular DNA digested with a restriction enzyme) can be used as the template DNA for PUREfrex® in case it contains at least the sequence described in the following section. Alternatively, mRNA can be also used to synthesize protein with PUREfrex®.
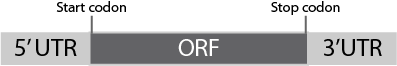
5' UTR
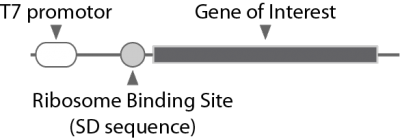
The template DNA must contain T7 promoter sequence and ribosome binding site (SD sequence) upstream of the gene encoding the protein of interest (Figure).
Stop Codon
PUREfrex® can be used with all stop codons. This is because the kit contains two release factors (RF1 and RF2) that recognize any of the three stop codons (TAA, ochre; TAG, amber; TGA, opal).
| Codon | Release factors |
|
|---|---|---|
| DNA | RNA | |
| TAG | UAG | RF1 |
| TAA | UAA | RF1, RF2 |
| TGA |
UGA | RF2 |
3' UTR
When using circular DNA, T7 terminator sequence to terminate transcription must be placed downstream of the gene encoding the protein of interest.
When using linear DNA, add at least 10 nucleotides downstream from stop codon. See “Examples of primers used for 2-step PCR” for an example of nucleotide sequence to add. T7 terminator sequence is not necessary when linear DNA is used.
Dissolving the template DNA
EDTA contained in TE buffer may reduce the efficiency of protein synthesis because it chelates magnesium ion necessary for transcription and translation reaction. We recommend dissolving the template DNA in EDTA-free buffer or Milli-Q water. Since contaminating RNase digests RNA (transcription products and tRNAs) in the reaction mixture, we recommend using only nuclease-free water, reagents, and instruments and wearing gloves and a mask.
Design of Template DNA
Certain nucleotide or amino acid sequence may reduce the yield of the protein of interest. In such cases, optimizing the sequence may improve the yield. Ribosomes, tRNAs, and translation factors in PUREfrex® are derived from E. coli. Therefore, in principle, it is recommended to design the gene encoding the protein of interest with codons optimized for translation in E. coli. When designing the template DNA sequence for PUREfrex®, please consider the following points, including the codons to be used.
For more information, please visit this site.
"6 Tips about the template DNA design"
ORF (Open Reading Frame)
Codon usage
Nucleotide sequences optimized for E. coli by available optimization tool may use single codon frequently used in E. coli. If the codon usage is excessively imbalanced in the optimized sequence, reassign properly according to the codon usage in the E. coli genome. The codon usage in the E. coli genome (W3110) is referred to the following website:
https://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=316407
Sequence that cause frameshift
Nucleotide sequences that are likely to cause frameshift during elongation, such as X/XXY/YYZ, should be replaced with other codons. A nucleotide sequence of “A/AAA/AAA” for 2 consecutive lysine residues, for example, should be replaced with “A/AAG/AAA” to avoid frameshift.
N-terminus
AT content just after the start codon
Select codons to maximize AT content in the region immediately after the start codon (usually up to 6th codon). In this region, use AT-rich codons over codons frequently used in E. coli.
Amino acid sequence at N-terminus
A low yield may occur if the second and third amino acids right after the first methionine are proline or glycine. Avoid using proline and glycine in this region if possible.
5' UTR
Ribosome Binding Site (SD sequence)
Without the SD sequence in the template DNA, the amount of protein synthesis is significantly reduced.
AT-rich region
The AT-rich region*1 upstream of the SD sequence should be at least 15 bases to increase the efficiency of translation.
- AT-rich region binds S1 protein of 30S ribosome subunit and increases translation efficiency.
Preparation of Template DNA
PCR product
PCR product purity
If bands other than that of the desired product are detected in electrophoresis after PCR, modify the PCR condition to reduce byproducts. The purity of the PCR product affect the efficiency of protein synthesis because proteins can be also synthesized from PCR-byproducts.
If byproducts are unable to eliminate by changing the PCR conditions, excise the target band from the gel and purify the product. Excise the band from the gel without using ultraviolet light, which can damage DNA (i.e., impair transcription). Blue light may be used, but exposure time should be kept to a minimum.
How much PCR product to add
The concentration of the template DNA added to the reaction mixture of PUREfrex® is defined as the number of DNA molecules (molar concentration) and it is added at a final concentration around 2 nM. About 2 nM is approximately 0.5 to 3 ng per 1 kbp for 1 µL of the reaction mixture.
Although unpurified PCR reaction can be directly used with PUREfrex®, do not add more than 10% (v/v) to the reaction mixture of PUREfrex®. The efficiency of both transcription and translation reaction may decrease when the salt concentration is altered by carryover from the PCR mixture. If the concentration of the PCR product is low, prepare DNA solution with a sufficient concentration using a DNA purification kit instead of adding more unpurified PCR mixture to the reaction mixture of PUREfrex®.
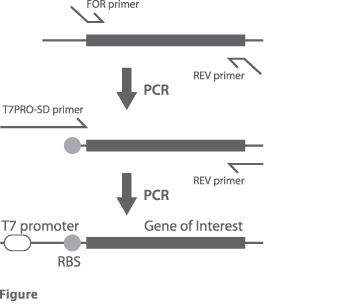
Overview of preparing the template DNA by 2-step PCR
Two-step PCR for preparing the template DNA is summarized in Figure. Use a high fidelity PCR enzyme (e.g., KOD (Toyobo), PrimeSTAR (TakaraBio), Pfu (Promega)) to prepare the template DNA. At the first step, use FOR primer and REV primer to amplify fragments containing a sequence from the ribosome binding site (RBS (SD sequence)) upstream of the gene encoding the protein of interest to a stop codon and at least 10 nucleotides. Adjust the PCR condition so that even minimal byproducts are produced at 1st PCR. At the second step, use T7PRO-SD primer and REV primer to amplify PCR fragments containing T7 promoter sequence.
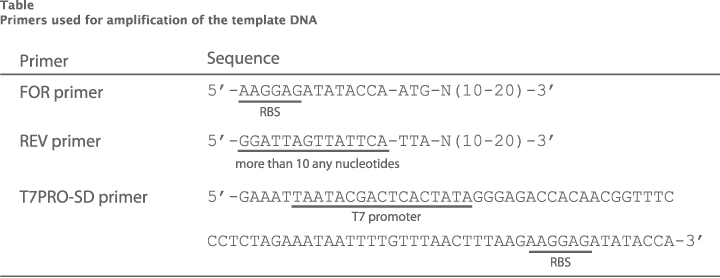
Examples of primers used for 2-step PCR
Primer sequences used for 2-step PCR are shown in Table.
The following site has a text sequence for the ORF of DHFR that also includes the 5'UTR and 3'TUR sequences
"Sequence of DHFR DNA"
Plasmid DNA
Usable vectors
Vectors that contain T7 promoter, SD sequence, and T7 terminator can be used such as pET vectors (Merck) and pQE vectors (Qiagen). The presence of the lac operator sequence may decrease the protein yield. We recommend using vectors without the lac operator sequence (e.g., pET17).
Plasmid DNA preparation
When preparing plasmid DNA, make sure that the activity of RNase used for purification is not contaminated in the final purified product. Using a filter-based purification kit such as QIAprep Spin Miniprep Kit (Qiagen) or Wizard Plus SV Minipreps DNA Purification System (Promega), for example, results in contamination of the final purified DNA solution by the RNase A contained in Lysis buffer. If adding this solution without further treatment to the PUREfrex® reaction mixture as template DNA, transcription products and other RNA in the reaction mixture are digested, inhibiting protein synthesis. Therefore, treat with phenol/chloroform to denature RNase and then re-purify by ethanol precipitation to prepare a DNA solution free of RNase activity. Adding RNase inhibitor to the PUREfrex® reaction mixture is also effective.
Plasmid DNA purified with Plasmid Mini Kit (Qiagen) has minimal contamination with RNase because the plasmid DNA eluted from resin is precipitated by adding isopropanol. Plasmid DNA purified with this kit are verified to use without any treatment.
How much plasmid DNA to add
The concentration of the template DNA added to the reaction mixture of PUREfrex® is defined as the number of DNA molecules (molar concentration) and it is added at a final concentration around 2 nM. About 2 nM is approximately 0.5 to 3 ng per 1 kbp for 1 µL of the reaction mixture. A 6 kbp plasmid DNA, for example, is be added at (0.5-3)×6 = 3-18 ng/µL, regardless of the length of the ORF.
RNA
In case using mRNA to synthesize the protein of interest, make sure that the mRNA contains SD sequence upstream of start codon. Add mRNA usually at a concentration of 0.1 to 1 µM to the reaction mixture. Since the optimal concentration depend on the sequence and purity of the mRNA, we recommend examining the optimal concentration in reference to this concentration range.
Request DNA design for PUREfrex®
<Consultation is free of charge!>
Send us the amino acid sequence of your target protein by the link below, and we will return a suitable DNA sequence based on our algorism considering important points for protein expression using PUREfrex®, such as codon optimization for whole ORF and N-terminus, UTRs, etc.